Release 0.6.1¶
Statsmodels 0.6.1 is a bugfix release. All users are encouraged to upgrade to 0.6.1.
See the list of fixed issues for specific backported fixes.
Release 0.6.0¶
Statsmodels 0.6.0 is another large release. It is the result of the work of 37 authors over the last year and includes over 1500 commits. It contains many new features, improvements, and bug fixes detailed below.
See the list of fixed issues for specific closed issues.
The following major new features appear in this version.
Generalized Estimating Equations¶
Generalized Estimating Equations (GEE) provide an approach to handling dependent data in a regression analysis. Dependent data arise commonly in practice, such as in a longitudinal study where repeated observations are collected on subjects. GEE can be viewed as an extension of the generalized linear modeling (GLM) framework to the dependent data setting. The familiar GLM families such as the Gaussian, Poisson, and logistic families can be used to accommodate dependent variables with various distributions.
Here is an example of GEE Poisson regression in a data set with four count-type repeated measures per subject, and three explanatory covariates.
import numpy as np
import statsmodels.api as sm
import statsmodels.formula.api as smf
data = sm.datasets.get_rdataset("epil", "MASS").data
md = smf.gee("y ~ age + trt + base", "subject", data,
cov_struct=sm.cov_struct.Independence(),
family=sm.families.Poisson())
mdf = md.fit()
print mdf.summary()
The dependence structure in a GEE is treated as a nuisance parameter and is modeled in terms of a “working dependence structure”. The statsmodels GEE implementation currently includes five working dependence structures (independent, exchangeable, autoregressive, nested, and a global odds ratio for working with categorical data). Since the GEE estimates are not maximum likelihood estimates, alternative approaches to some common inference procedures have been developed. The statsmodels GEE implementation currently provides standard errors, Wald tests, score tests for arbitrary parameter contrasts, and estimates and tests for marginal effects. Several forms of standard errors are provided, including robust standard errors that are approximately correct even if the working dependence structure is misspecified.
Seasonality Plots¶
Adding functionality to look at seasonality in plots. Two new functions are sm.graphics.tsa.month_plot and sm.graphics.tsa.quarter_plot. Another function sm.graphics.tsa.seasonal_plot is available for power users.
import statsmodels.api as sm
import pandas as pd
dta = sm.datasets.elnino.load_pandas().data
dta['YEAR'] = dta.YEAR.astype(int).astype(str)
dta = dta.set_index('YEAR').T.unstack()
dates = map(lambda x : pd.datetools.parse('1 '+' '.join(x)),
dta.index.values)
dta.index = pd.DatetimeIndex(dates, freq='M')
fig = sm.tsa.graphics.month_plot(dta)
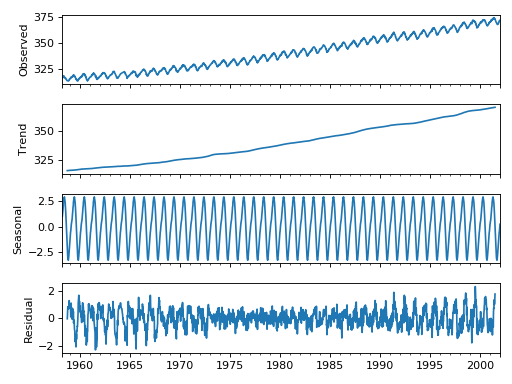
Seasonal Decomposition¶
We added a naive seasonal decomposition tool in the same vein as R’s decompose. This function can be found as sm.tsa.seasonal_decompose.
import statsmodels.api as sm
dta = sm.datasets.co2.load_pandas().data
# deal with missing values. see issue
dta.co2.interpolate(inplace=True)
res = sm.tsa.seasonal_decompose(dta.co2)
res.plot()
(Source code, png, hires.png, pdf)
Addition of Linear Mixed Effects Models (MixedLM)
Linear Mixed Effects Models¶
Linear Mixed Effects models are used for regression analyses involving dependent data. Such data arise when working with longitudinal and other study designs in which multiple observations are made on each subject. Two specific mixed effects models are “random intercepts models”, where all responses in a single group are additively shifted by a value that is specific to the group, and “random slopes models”, where the values follow a mean trajectory that is linear in observed covariates, with both the slopes and intercept being specific to the group. The Statsmodels MixedLM implementation allows arbitrary random effects design matrices to be specified for the groups, so these and other types of random effects models can all be fit.
Here is an example of fitting a random intercepts model to data from a longitudinal study:
import statsmodels.api as sm
import statsmodels.formula.api as smf
data = sm.datasets.get_rdataset('dietox', 'geepack', cache=True).data
md = smf.mixedlm("Weight ~ Time", data, groups=data["Pig"])
mdf = md.fit()
print mdf.summary()
The Statsmodels LME framework currently supports post-estimation inference via Wald tests and confidence intervals on the coefficients, profile likelihood analysis, likelihood ratio testing, and AIC. Some limitations of the current implementation are that it does not support structure more complex on the residual errors (they are always homoscedastic), and it does not support crossed random effects. We hope to implement these features for the next release.
Wrapping X-12-ARIMA/X-13-ARIMA¶
It is now possible to call out to X-12-ARIMA or X-13ARIMA-SEATS from statsmodels. These libraries must be installed separately.
import statsmodels.api as sm
dta = sm.datasets.co2.load_pandas().data
dta.co2.interpolate(inplace=True)
dta = dta.resample('M').last()
res = sm.tsa.x13_arima_select_order(dta.co2)
print(res.order, res.sorder)
results = sm.tsa.x13_arima_analysis(dta.co2)
fig = results.plot()
fig.set_size_inches(12, 5)
fig.tight_layout()
Other important new features¶
The AR(I)MA models now have a
plot_predictmethod to plot forecasts and confidence intervals.The Kalman filter Cython code underlying AR(I)MA estimation has been substantially optimized. You can expect speed-ups of one to two orders of magnitude.
Added
sm.tsa.arma_order_select_ic. A convenience function to quickly get the information criteria for use in tentative order selection of ARMA processes.Plotting functions for timeseries is now imported under the
sm.tsa.graphicsnamespace in addition tosm.graphics.tsa.New distributions.ExpandedNormal class implements the Edgeworth expansion for weakly non-normal distributions.
New datasets: Added new datasets for examples.
sm.datasets.co2is a univariate time-series dataset of weekly co2 readings. It exhibits a trend and seasonality and has missing values.Added robust skewness and kurtosis estimators in
sm.stats.stattools.robust_skewnessandsm.stats.stattools.robust_kurtosis, respectively. An alternative robust measure of skewness has been added insm.stats.stattools.medcouple.New functions added to correlation tools: corr_nearest_factor finds the closest factor-structured correlation matrix to a given square matrix in the Frobenius norm; corr_thresholded efficiently constructs a hard-thresholded correlation matrix using sparse matrix operations.
New dot_plot in graphics: A dotplot is a way to visualize a small dataset in a way that immediately conveys the identity of every point in the plot. Dotplots are commonly seen in meta-analyses, where they are known as “forest plots”, but can be used in many other settings as well. Most tables that appear in research papers can be represented graphically as a dotplot.
Statsmodels has added custom warnings to
statsmodels.tools.sm_exceptions. By default all of these warnings will be raised whenever appropriate. Usewarnings.simplefilterto turn them off, if desired.Allow control over the namespace used to evaluate formulas with patsy via the
eval_envkeyword argument. See the Namespaces documentation for more information.
Major Bugs fixed¶
Backwards incompatible changes and deprecations¶
RegressionResults.norm_resid is now a readonly property, rather than a function.
The function
statsmodels.tsa.filters.arfilterhas been removed. This did not compute a recursive AR filter but was instead a convolution filter. Two new functions have been added with clearer namessm.tsa.filters.recursive_filterandsm.tsa.filters.convolution_filter.
Development summary and credits¶
The previous version (0.5.0) was released August 14, 2014. Since then we have closed a total of 528 issues, 276 pull requests, and 252 regular issues. Refer to the detailed list for more information.
This release is a result of the work of the following 37 authors who contributed a total of 1531 commits. If for any reason we have failed to list your name in the below, please contact us:
A blurb about the number of changes and the contributors list.
Alex Griffing <argriffi-at-ncsu.edu>
Alex Parij <paris.alex-at-gmail.com>
Ana Martinez Pardo <anamartinezpardo-at-gmail.com>
Andrew Clegg <andrewclegg-at-users.noreply.github.com>
Ben Duffield <bduffield-at-palantir.com>
Chad Fulton <chad-at-chadfulton.com>
Chris Kerr <cjk34-at-cam.ac.uk>
Eric Chiang <eric.chiang.m-at-gmail.com>
Evgeni Burovski <evgeni-at-burovski.me>
gliptak <gliptak-at-gmail.com>
Hans-Martin von Gaudecker <hmgaudecker-at-uni-bonn.de>
Jan Schulz <jasc-at-gmx.net>
jfoo <jcjf1983-at-gmail.com>
Joe Hand <joe.a.hand-at-gmail.com>
Josef Perktold <josef.pktd-at-gmail.com>
jsphon <jonathanhon-at-hotmail.com>
Justin Grana <jg3705a-at-student.american.edu>
Kerby Shedden <kshedden-at-umich.edu>
Kevin Sheppard <kevin.sheppard-at-economics.ox.ac.uk>
Kyle Beauchamp <kyleabeauchamp-at-gmail.com>
Lars Buitinck <l.buitinck-at-esciencecenter.nl>
Max Linke <max_linke-at-gmx.de>
Miroslav Batchkarov <mbatchkarov-at-gmail.com>
m <mngu2382-at-gmail.com>
Padarn Wilson <padarn-at-gmail.com>
Paul Hobson <pmhobson-at-gmail.com>
Pietro Battiston <me-at-pietrobattiston.it>
Radim Řehůřek <radimrehurek-at-seznam.cz>
Ralf Gommers <ralf.gommers-at-googlemail.com>
Richard T. Guy <richardtguy84-at-gmail.com>
Roy Hyunjin Han <rhh-at-crosscompute.com>
Skipper Seabold <jsseabold-at-gmail.com>
Tom Augspurger <thomas-augspurger-at-uiowa.edu>
Trent Hauck <trent.hauck-at-gmail.com>
Valentin Haenel <valentin.haenel-at-gmx.de>
Vincent Arel-Bundock <varel-at-umich.edu>
Yaroslav Halchenko <debian-at-onerussian.com>
Note
Obtained by running git log v0.5.0..HEAD --format='* %aN <%aE>' | sed 's/@/\-at\-/' | sed 's/<>//' | sort -u.
Issues closed in the 0.6.0 development cycle¶
Issues closed in 0.6.0¶
GitHub stats for 2013/08/14 - 2014/10/15 (tag: v0.5.0)
We closed a total of 528 issues, 276 pull requests and 252 regular issues;
this is the full list (generated with the script tools/github_stats.py):
This list is automatically generated and may be incomplete.
Pull Requests (276):
PR #2044: ENH: Allow unit interval for binary models. Closes #2040.
PR #1426: ENH: Import arima_process stuff into tsa.api
PR #2042: Fix two minor typos in contrast.py
PR #2034: ENH: Handle missing for extra data with formulas
PR #2035: MAINT: Remove deprecated code for 0.6
PR #1325: ENH: add the Edgeworth expansion based on the normal distribution
PR #2032: DOC: What it is what it is.
PR #2031: ENH: Expose patsy eval_env to users.
PR #2028: ENH: Fix numerical issues in links and families.
PR #2029: DOC: Fix versions to match other docs.
PR #1647: ENH: Warn on non-convergence.
PR #2014: BUG: Fix forecasting for ARIMA with d == 2
PR #2013: ENH: Better error message on object dtype
PR #2012: BUG: 2d 1 columns -> 1d. Closes #322.
PR #2009: DOC: Update after refactor. Use code block.
PR #2008: ENH: Add wrapper for MixedLM
PR #1954: ENH: PHReg formula improvements
PR #2007: BLD: Fix build issues
PR #2006: BLD: Do not generate cython on clean. Closes #1852.
PR #2000: BLD: Let pip/setuptools handle dependencies that aren’t installed at all.
PR #1999: Gee offset exposure 1994 rebased
PR #1998: BUG/ENH Lasso emptymodel rebased
PR #1989: BUG/ENH: WLS generic robust cov_type didn’t use whitened,
PR #1587: ENH: Wrap X12/X13-ARIMA AUTOMDL. Closes #442.
PR #1563: ENH: Add plot_predict method to ARIMA models.
PR #1995: BUG: Fix issue #1993
PR #1981: ENH: Add api for covstruct. Clear __init__. Closes #1917.
PR #1996: DEV: Ignore .venv file.
PR #1982: REF: Rename jac -> score_obs. Closes #1785.
PR #1987: BUG tsa pacf, base bootstrap
PR #1986: Bug multicomp 1927 rebased
PR #1984: Docs add gee.rst
PR #1985: Bug uncentered latex table 1929 rebased
PR #1983: BUG: Fix compat asunicode
PR #1574: DOC: Fix math.
PR #1980: DOC: Documentation fixes
PR #1974: REF/Doc beanplot change default color, add notebook
PR #1978: ENH: Check input to binary models
PR #1979: BUG: Typo
PR #1976: ENH: Add _repr_html_ to SimpleTable
PR #1977: BUG: Fix import refactor victim.
PR #1975: BUG: Yule walker cast to float
PR #1973: REF: Move and expose webuse
PR #1972: TST: Add testing against NumPy 1.9 and matplotlib 1.4
PR #1939: ENH: Binstar build files
PR #1952: REF/DOC: Misc
PR #1940: REF: refactor and speedup of mixed LME
PR #1937: ENH: Quick access to online documentation
PR #1942: DOC: Rename Change README type to rst
PR #1938: ENH: Enable Python 3.4 testing
PR #1924: Bug gee cov type 1906 rebased
PR #1870: robust covariance, cov_type in fit
PR #1859: BUG: Don’t use negative indexing with k_ar == 0. Closes #1858.
PR #1914: BUG: LikelihoodModelResults.pvalues use df_resid_inference
PR #1899: TST: fix assert_equal for pandas index
PR #1895: Bug multicomp pandas
PR #1894: BUG fix more ix indexing cases for pandas compat
PR #1889: BUG: fix ytick positions closes #1561
PR #1887: Bug pandas compat asserts
PR #1888: TST test_corrpsd Test_Factor: add noise to data
PR #1886: BUG pandas 0.15 compatibility in grouputils labels
PR #1885: TST: corr_nearest_factor, more informative tests
PR #1884: Fix: Add compat code for pd.Categorical in pandas>=0.15
PR #1883: BUG: add _ctor_param to TransfGen distributions
PR #1872: TST: fix _infer_freq for pandas .14+ compat
PR #1867: Ref covtype fit
PR #1865: Disable tst distribution 1864
PR #1856: _spg_optim returns history of objective function values
PR #1854: BLD: Don’t hard-code path for building notebooks. Closes #1249
PR #1851: MAINT: Cor nearest factor tests
PR #1847: Newton regularize
PR #1623: BUG Negbin fit regularized
PR #1797: BUG/ENH: fix and improve constant detection
PR #1770: TST: anova with -1 noconstant, add tests
PR #1837: Allow group variable to be passed as variable name when using formula
PR #1839: BUG: GEE score
PR #1830: BUG/ENH Use t
PR #1832: TST error with scipy 0.14 location distribution class
PR #1827: fit_regularized for linear models rebase 1674
PR #1825: Phreg 1312 rebased
PR #1826: Lme api docs
PR #1824: Lme profile 1695 rebased
PR #1823: Gee cat subclass 1694 rebase
PR #1781: ENH: Glm add score_obs
PR #1821: Glm maint #1734 rebased
PR #1820: BUG: revert change to conf_int in PR #1819
PR #1819: Docwork
PR #1772: REF: cov_params allow case of only cov_params_default is defined
PR #1771: REF numpy >1.9 compatibility, indexing into empty slice closes #1754
PR #1769: Fix ttest 1d
PR #1766: TST: TestProbitCG increase bound for fcalls closes #1690
PR #1709: BLD: Made build extensions more flexible
PR #1714: WIP: fit_constrained
PR #1706: REF: Use fixed params in test. Closes #910.
PR #1701: BUG: Fix faulty logic. Do not raise when missing=’raise’ and no missing data.
PR #1699: TST/ENH StandardizeTransform, reparameterize TestProbitCG
PR #1697: Fix for statsmodels/statsmodels#1689
PR #1692: OSL Example: redundant cell in example removed
PR #1688: Kshedden mixed rebased of #1398
PR #1629: Pull request to fix bandwidth bug in issue 597
PR #1666: Include pyx in sdist but don’t install
PR #1683: TST: GLM shorten random seed closes #1682
PR #1681: Dotplot kshedden rebased of 1294
PR #1679: BUG: Fix problems with predict handling offset and exposure
PR #1677: Update docstring of RegressionModel.predict()
PR #1635: Allow offset and exposure to be used together with log link; raise excep…
PR #1676: Tests for SVAR
PR #1671: ENH: avoid hard-listed bandwidths – use present dictionary (+typos fixed)
PR #1643: Allow matrix structure in covariance matrices to be exploited
PR #1657: BUG: Fix refactor victim.
PR #1630: DOC: typo, “interecept”
PR #1619: MAINT: Dataset docs cleanup and automatic build of docs
PR #1612: BUG/ENH Fix negbin exposure #1611
PR #1610: BUG/ENH fix llnull, extra kwds to recreate model
PR #1582: BUG: wls_prediction_std fix weight handling, see 987
PR #1613: BUG: Fix proportions allpairs #1493
PR #1607: TST: adjust precision, CI Debian, Ubuntu testing
PR #1603: ENH: Allow start_params in GLM
PR #1600: CLN: Regression plots fixes
PR #1592: DOC: Additions and fixes
PR #1520: CLN: Refactored so that there is no longer a need for 2to3
PR #1585: Cor nearest 1384 rebased
PR #1553: Gee maint 1528 rebased
PR #1583: BUG: For ARMA(0,0) ensure 1d bse and fix summary.
PR #1580: DOC: Fix links. [skip ci]
PR #1572: DOC: Fix link title [skip ci]
PR #1566: BLD: Fix copy paste path error for >= 3.3 Windows builds
PR #1524: ENH: Optimize Cython code. Use scipy blas function pointers.
PR #1560: ENH: Allow ARMA(0,0) in order selection
PR #1559: MAINT: Recover lost commits from vbench PR
PR #1554: Silenced test output introduced in medcouple
PR #1234: ENH: Robust skewness, kurtosis and medcouple measures
PR #1484: ENH: Add naive seasonal decomposition function
PR #1551: COMPAT: Fix failing test on Python 2.6
PR #1472: ENH: using human-readable group names instead of integer ids in MultiComparison
PR #1437: ENH: accept non-int definitions of cluster groups
PR #1550: Fix test gmm poisson
PR #1549: TST: Fix locally failing tests.
PR #1121: WIP: Refactor optimization code.
PR #1547: COMPAT: Correct bit_length for 2.6
PR #1545: MAINT: Fix missed usage of deprecated tools.rank
PR #1196: REF: ensure O(N log N) when using fft for acf
PR #1154: DOC: Add links for build machines.
PR #1546: DOC: Fix link to wrong notebook
PR #1383: MAINT: Deprecate rank in favor of np.linalg.matrix_rank
PR #1432: COMPAT: Add NumpyVersion from scipy
PR #1438: ENH: Option to avoid “center” environment.
PR #1544: BUG: Travis miniconda
PR #1510: CLN: Improve warnings to avoid generic warnings messages
PR #1543: TST: Suppress RuntimeWarning for L-BFGS-B
PR #1507: CLN: Silence test output
PR #1540: BUG: Correct derivative for exponential transform.
PR #1536: BUG: Restores coveralls for a single build
PR #1535: BUG: Fixes for 2.6 test failures, replacing astype(str) with apply(str)
PR #1523: Travis miniconda
PR #1533: DOC: Fix link to code on github
PR #1531: DOC: Fix stale links with linkcheck
PR #1530: DOC: Fix link
PR #1527: DOCS: Update docs add FAQ page
PR #1525: DOC: Update with Python 3.4 build notes
PR #1518: DOC: Ask for release notes and example.
PR #1516: DOC: Update examples contributing docs for current practice.
PR #1517: DOC: Be clear about data attribute of Datasets
PR #1515: DOC: Fix broken link
PR #1514: DOC: Fix formula import convention.
PR #1506: BUG: Format and decode errors in Python 2.6
PR #1505: TST: Test co2 load_data for Python 3.
PR #1504: BLD: New R versions require NAMESPACE file. Closes #1497.
PR #1483: ENH: Some utility functions for working with dates
PR #1482: REF: Prefer filters.api to __init__
PR #1481: ENH: Add weekly co2 dataset
PR #1474: DOC: Add plots for standard filter methods.
PR #1471: DOC: Fix import
PR #1470: DOC/BLD: Log code exceptions from nbgenerate
PR #1469: DOC: Fix bad links
PR #1468: MAINT: CSS fixes
PR #1463: DOC: Remove defunct argument. Change default kw. Closes #1462.
PR #1452: STY: import pandas as pd
PR #1458: BUG/BLD: exclude sandbox in relative path, not absolute
PR #1447: DOC: Only build and upload docs if we need to.
PR #1445: DOCS: Example landing page
PR #1436: DOC: Fix auto doc builds.
PR #1431: DOC: Add default for getenv. Fix paths. Add print_info
PR #1429: MAINT: Use ip_directive shipped with IPython
PR #1427: TST: Make tests fit quietly
PR #1424: ENH: Consistent results for transform_slices
PR #1421: ENH: Add grouping utilities code
PR #1419: Gee 1314 rebased
PR #1414: TST temporarily rename tests probplot other to skip them
PR #1403: Bug norm expan shapes
PR #1417: REF: Let subclasses keep kwds attached to data.
PR #1416: ENH: Make handle_data overwritable by subclasses.
PR #1410: ENH: Handle missing is none
PR #1402: REF: Expose missing data handling as classmethod
PR #1387: MAINT: Fix failing tests
PR #1406: MAINT: Tools improvements
PR #1404: Tst fix genmod link tests
PR #1396: REF: Multipletests reduce memory usage
PR #1380: DOC :Update vector_ar.rst
PR #1381: BLD: Don’t check dependencies on egg_info for pip. Closes #1267.
PR #1302: BUG: Fix typo.
PR #1375: STY: Remove unused imports and comment out unused libraries in setup.py
PR #1143: DOC: Update backport notes for new workflow.
PR #1374: ENH: Import tsaplots into tsa namespace. Closes #1359.
PR #1369: STY: Pep-8 cleanup
PR #1370: ENH: Support ARMA(0,0) models.
PR #1368: STY: Pep 8 cleanup
PR #1367: ENH: Make sure mle returns attach to results.
PR #1365: STY: Import and pep 8 cleanup
PR #1364: ENH: Get rid of hard-coded lbfgs. Closes #988.
PR #1363: BUG: Fix typo.
PR #1361: ENH: Attach mlefit to results not model.
PR #1360: ENH: Import adfuller into tsa namespace
PR #1346: STY: PEP-8 Cleanup
PR #1344: BUG: Use missing keyword given to ARMA.
PR #1340: ENH: Protect against ARMA convergence failures.
PR #1334: ENH: ARMA order select convenience function
PR #1339: Fix typos
PR #1336: REF: Get rid of plain assert.
PR #1333: STY: __all__ should be after imports.
PR #1332: ENH: Add Bunch object to tools.
PR #1331: ENH: Always use unicode.
PR #1329: BUG: Decode metadata to utf-8. Closes #1326.
PR #1330: DOC: Fix typo. Closes #1327.
PR #1185: Added support for pandas when pandas was installed directly from git trunk
PR #1315: MAINT: Change back to path for build box
PR #1305: TST: Update hard-coded path.
PR #1290: ENH: Add seasonal plotting.
PR #1296: BUG/TST: Fix ARMA forecast when start == len(endog). Closes #1295
PR #1292: DOC: cleanup examples folder and webpage
PR #1286: Make sure PeriodIndex passes through tsa. Closes #1285.
PR #1271: Silverman enhancement - Issue #1243
PR #1264: Doc work GEE, GMM, sphinx warnings
PR #1179: REF/TST: ProbPlot now uses resettable_cache and added some kwargs to plotting fxns
PR #1225: Sandwich mle
PR #1258: Gmm new rebased
PR #1255: ENH add GEE to genmod
PR #1254: REF: Results.predict convert to array and adjust shape
PR #1192: TST: enable tests for llf after change to WLS.loglike see #1170
PR #1253: Wls llf fix
PR #1233: sandbox kernels bugs uniform kernel and confint
PR #1240: Kde weights 1103 823
PR #1228: Add default value tags to adfuller() docs
PR #1198: fix typo
PR #1230: BUG: numerical precision in resid_pearson with perfect fit #1229
PR #1214: Compare lr test rebased
PR #1200: BLD: do not install *.pyx *.c MANIFEST.in
PR #1202: MAINT: Sort backports to make applying easier.
PR #1157: Tst precision master
PR #1161: add a fitting interface for simultaneous log likelihood and score, for lbfgs, tested with MNLogit
PR #1160: DOC: update scipy version from 0.7 to 0.9.0
PR #1147: ENH: add lbfgs for fitting
PR #1156: ENH: Raise on 0,0 order models in AR(I)MA. Closes #1123
PR #1149: BUG: Fix small data issues for ARIMA.
PR #1092: Fixed duplicate svd in RegressionModel
PR #1139: TST: Silence tests
PR #1135: Misc style
PR #1088: ENH: add predict_prob to poisson
PR #1125: REF/BUG: Some GLM cleanup. Used trimmed results in NegativeBinomial variance.
PR #1124: BUG: Fix ARIMA prediction when fit without a trend.
PR #1118: DOC: Update gettingstarted.rst
PR #1117: Update ex_arma2.py
PR #1107: REF: Deprecate stand_mad. Add center keyword to mad. Closes #658.
PR #1089: ENH: exp(poisson.logpmf()) for poisson better behaved.
PR #1077: BUG: Allow 1d exog in ARMAX forecasting.
PR #1075: BLD: Fix build issue on some versions of easy_install.
PR #1071: Update setup.py to fix broken install on OSX
PR #1052: DOC: Updating contributing docs
PR #1136: RLS: Add IPython tools for easier backporting of issues.
PR #1091: DOC: minor git typo
PR #1082: coveralls support
PR #1072: notebook examples title cell
PR #1056: Example: reg diagnostics
PR #1057: COMPAT: Fix py3 caching for get_rdatasets.
PR #1045: DOC/BLD: Update from nbconvert to IPython 1.0.
PR #1026: DOC/BLD: Add LD_LIBRARY_PATH to env for docs build.
Issues (252):
#2040: enh: fractional Logit, Probit
#1220: missing in extra data (example sandwiches, robust covariances)
#1877: error with GEE on missing data.
#805: nan with categorical in formula
#2036: test in links require exact class so Logit can’t work in place of logit
#2010: Go over deprecations again for 0.6.
#1303: patsy library not automatically installed
#2024: genmod Links numerical improvements
#2025: GEE requires exact import for cov_struct
#2017: Matplotlib warning about too many figures
#724: check warnings
#1562: ARIMA forecasts are hard-coded for d=1
#880: DataFrame with bool type not cast correctly.
#1992: MixedLM style
#322: acf / pacf do not work on pandas objects
#1317: AssertionError: attr is not equal [dtype]: dtype(‘object’) != dtype(‘datetime64[ns]’)
#1875: dtype bug object arrays (raises in clustered standard errors code)
#1842: dtype object, glm.fit() gives AttributeError: sqrt
#1300: Doc errors, missing
#1164: RLM cov_params, t_test, f_test don’t use bcov_scaled
#1019: 0.6.0 Roadmap
#554: Prediction Standard Errors
#333: ENH tools: squeeze in R export file
#1990: MixedLM does not have a wrapper
#1897: Consider depending on setuptools in setup.py
#2003: pip install now fails silently
#1852: do not cythonize when cleaning up
#1991: GEE formula interface does not take offset/exposure
#442: Wrap x-12 arima
#1993: MixedLM bug
#1917: API: GEE access to genmod.covariance_structure through api
#1785: REF: rename jac -> score_obs
#1969: pacf has incorrect standard errors for lag 0
#1434: A small bug in GenericLikelihoodModelResults.bootstrap()
#1408: BUG test failure with tsa_plots
#1337: DOC: HCCM are now available for WLS
#546: influence and outlier documentation
#1532: DOC: Related page is out of date
#1386: Add minimum matplotlib to docs
#1068: DOC: keeping documentation of old versions on sourceforge
#329: link to examples and datasets from module pages
#1804: PDF documentation for statsmodels
#202: Extend robust standard errors for WLS/GLS
#1519: Link to user-contributed examples in docs
#1053: inconvenient: logit when endog is (1,2) instead of (0,1)
#1555: SimpleTable: add repr html for ipython notebook
#1366: Change default start_params to .1 in ARMA
#1869: yule_walker (from statsmodels.regression) raises exception when given an integer array
#1651: statsmodels.tsa.ar_model.ARResults.predict
#1738: GLM robust sandwich covariance matrices
#1779: Some directories under statsmodels dont have __init_.py
#1242: No support for (0, 1, 0) ARIMA Models
#1571: expose webuse, use cache
#1860: ENH/BUG/DOC: Bean plot should allow for separate widths of bean and violins.
#1831: TestRegressionNM.test_ci_beta2 i386 AssertionError
#1079: bugfix release 0.5.1
#1338: Raise Warning for HCCM use in WLS/GLS
#1430: scipy min version / issue
#276: memoize, last argument wins, how to attach sandwich to Results?
#1943: REF/ENH: LikelihoodModel.fit optimization, make hessian optional
#1957: BUG: Re-create OLS model using _init_keys
#1905: Docs: online docs are missing GEE
#1898: add python 3.4 to continuous integration testing
#1684: BUG: GLM NegativeBinomial: llf ignores offset and exposure
#1256: REF: GEE handling of default covariance matrices
#1760: Changing covariance_type on results
#1906: BUG: GEE default covariance is not used
#1931: BUG: GEE subclasses NominalGEE don’t work with pandas exog
#1904: GEE Results doesn’t have a Wrapper
#1918: GEE: required attributes missing, df_resid
#1919: BUG GEE.predict uses link instead of link.inverse
#1858: BUG: arimax forecast should special case k_ar == 0
#1903: BUG: pvalues for cluster robust, with use_t don’t use df_resid_inference
#1243: kde silverman bandwidth for non-gaussian kernels
#1866: Pip dependencies
#1850: TST test_corr_nearest_factor fails on Ubuntu
#292: python 3 examples
#1868: ImportError: No module named compat [ from statsmodels.compat import lmap ]
#1890: BUG tukeyhsd nan in group labels
#1891: TST test_gmm outdated pandas, compat
#1561: BUG plot for tukeyhsd, MultipleComparison
#1864: test failure sandbox distribution transformation with scipy 0.14.0
#576: Add contributing guidelines
#1873: GenericLikelihoodModel is not picklable
#1822: TST failure on Ubuntu pandas 0.14.0 , problems with frequency
#1249: Source directory problem for notebook examples
#1855: anova_lm throws error on models created from api.ols but not formula.api.ols
#1853: a large number of hardcoded paths
#1792: R² adjusted strange after including interaction term
#1794: REF: has_constant, k_constant, include implicit constant detection in base
#1454: NegativeBinomial missing fit_regularized method
#1615: REF DRYing fit methods
#1453: Discrete NegativeBinomialModel regularized_fit ValueError: matrices are not aligned
#1836: BUG Got an TypeError trying to import statsmodels.api
#1829: BUG: GLM summary show “t” use_t=True for summary
#1828: BUG summary2 doesn’t propagate/use use_t
#1812: BUG/ REF conf_int and use_t
#1835: Problems with installation using easy_install
#1801: BUG ‘f_gen’ missing in scipy 0.14.0
#1803: Error revealed by numpy 1.9.0r1
#1834: stackloss
#1728: GLM.fit maxiter=0 incorrect
#1795: singular design with offset ?
#1730: ENH/Bug cov_params, generalize, avoid ValueError
#1754: BUG/REF: assignment to slices in numpy >= 1.9 (emplike)
#1409: GEE test errors on Debian Wheezy
#1521: ubuntu failues: tsa_plot and grouputils
#1415: test failure test_arima.test_small_data
#1213: df_diff in anova_lm
#1323: Contrast Results after t_test summary broken for 1 parameter
#109: TestProbitCG failure on Ubuntu
#1690: TestProbitCG: 8 failing tests (Python 3.4 / Ubuntu 12.04)
#1763: Johansen method doesn’t give correct index values
#1761: doc build failures: ipython version ? ipython directive
#1762: Unable to build
#1745: UnicodeDecodeError raised by get_rdataset(“Guerry”, “HistData”)
#611: test failure foreign with pandas 0.7.3
#1700: faulty logic in missing handling
#1648: ProbitCG failures
#1689: test_arima.test_small_data: SVD fails to converge (Python 3.4 / Ubuntu 12.04)
#597: BUG: nonparametric: kernel, efficient=True changes bw even if given
#1606: BUILD from sdist broken if cython available
#1246: test failure test_anova.TestAnova2.test_results
#50: t_test, f_test, model.py for normal instead of t-distribution
#1655: newey-west different than R?
#1682: TST test failure on Ubuntu, random.seed
#1614: docstring for regression.linear_model.RegressionModel.predict() does not match implementation
#1318: GEE and GLM scale parameter
#519: L1 fit_regularized cleanup, comments
#651: add structure to example page
#1067: Kalman Filter convergence. How close is close enough?
#1281: Newton convergence failure prints warnings instead of warning
#1628: Unable to install statsmodels in the same requirements file as numpy, pandas, etc.
#617: Problem in installing statsmodel in Fedora 17 64-bit
#935: ll_null in likelihoodmodels discrete
#704: datasets.sunspot: wrong link in description
#1222: NegativeBinomial ignores exposure
#1611: BUG NegativeBinomial ignores exposure and offset
#1608: BUG: NegativeBinomial, llnul is always default ‘nb2’
#1221: llnull with exposure ?
#1493: statsmodels.stats.proportion.proportions_chisquare_allpairs has hardcoded value
#1260: GEE test failure on Debian
#1261: test failure on Debian
#443: GLM.fit does not allow start_params
#1602: Fitting GLM with a pre-assigned starting parameter
#1601: Fitting GLM with a pre-assigned starting parameter
#890: regression_plots problems (pylint) and missing test coverage
#1598: Is “old” string formatting Python 3 compatible?
#1589: AR vs ARMA order specification
#1134: Mark knownfails
#1259: Parameterless models
#616: python 2.6, python 3 in single codebase
#1586: Kalman Filter errors with new pyx
#1565: build_win_bdist*_py3*.bat are using the wrong compiler
#843: UnboundLocalError When trying to install OS X
#713: arima.fit performance
#367: unable to install on RHEL 5.6
#1548: testtransf error
#1478: is sm.tsa.filters.arfilter an AR filter?
#1420: GMM poisson test failures
#1145: test_multi noise
#1539: NegativeBinomial strange results with bfgs
#936: vbench for statsmodels
#1153: Where are all our testing machines?
#1500: Use Miniconda for test builds
#1526: Out of date docs
#1311: BUG/BLD 3.4 compatibility of cython c files
#1513: build on osx -python-3.4
#1497: r2nparray needs NAMESPACE file
#1502: coveralls coverage report for files is broken
#1501: pandas in/out in predict
#1494: truncated violin plots
#1443: Crash from python.exe using linear regression of statsmodels
#1462: qqplot line kwarg is broken/docstring is wrong
#1457: BUG/BLD: Failed build if “sandbox” anywhere in statsmodels path
#1441: wls function: syntax error “unexpected EOF while parsing” occurs when name of dependent variable starts with digits
#1428: ipython_directive doesn’t work with ipython master
#1385: SimpleTable in Summary (e.g. OLS) is slow for large models
#1399: UnboundLocalError: local variable ‘fittedvalues’ referenced before assignment
#1377: TestAnova2.test_results fails with pandas 0.13.1
#1394: multipletests: reducing memory consumption
#1267: Packages cannot have both pandas and statsmodels in install_requires
#1359: move graphics.tsa to tsa.graphics
#356: docs take up a lot of space
#988: AR.fit no precision options for fmin_l_bfgs_b
#990: AR fit with bfgs: large score
#14: arma with exog
#1348: reset_index + set_index with drop=False
#1343: ARMA doesn’t pass missing keyword up to TimeSeriesModel
#1326: formula example notebook broken
#1327: typo in docu-code for “Outlier and Influence Diagnostic Measures”
#1309: Box-Cox transform (some code needed: lambda estimator)
#1059: sm.tsa.ARMA making ma invertibility
#1295: Bug in ARIMA forecasting when start is int len(endog) and dates are given
#1285: tsa models fail on PeriodIndex with pandas
#1269: KPSS test for stationary processes
#1268: Feature request: Exponential smoothing
#1250: DOCs error in var_plots
#1032: Poisson predict breaks on list
#347: minimum number of observations - document or check ?
#1170: WLS log likelihood, aic and bic
#1187: sm.tsa.acovf fails when both unbiased and fft are True
#1239: sandbox kernels, problems with inDomain
#1231: sandbox kernels confint missing alpha
#1245: kernels cosine differs from Stata
#823: KDEUnivariate with weights
#1229: precision problems in degenerate case
#1219: select_order
#1206: REF: RegressionResults cov-HCx into cached attributes
#1152: statsmodels failing tests with pandas master
#1195: pyximport.install() before import api crash
#1066: gmm.IV2SLS has wrong predict signature
#1186: OLS when exog is 1d
#1113: TST: precision too high in test_normality
#1159: scipy version is still >= 0.7?
#1108: SyntaxError: unqualified exec is not allowed in function ‘test_EvalEnvironment_capture_flag
#1116: Typo in Example Doc?
#1123: BUG : arima_model._get_predict_out_of_sample, ignores exogenous of there is no trend ?
#1155: ARIMA - The computed initial AR coefficients are not stationary
#979: Win64 binary can’t find Python installation
#1046: TST: test_arima_small_data_bug on current master
#1146: ARIMA fit failing for small set of data due to invalid maxlag
#1081: streamline linear algebra for linear model
#1138: BUG: pacf_yw doesn’t demean
#1127: Allow linear link model with Binomial families
#1122: no data cleaning for statsmodels.genmod.families.varfuncs.NegativeBinomial()
#658: robust.mad is not being computed correctly or is non-standard definition; it returns the median
#1076: Some issues with ARMAX forecasting
#1073: easy_install sandbox violation
#1115: EasyInstall Problem
#1106: bug in robust.scale.mad?
#1102: Installation Problem
#1084: DataFrame.sort_index does not use ascending when then value is a list with a single element
#393: marginal effects in discrete choice do not have standard errors defined
#1078: Use pandas.version.short_version
#96: deepcopy breaks on ResettableCache
#1055: datasets.get_rdataset string decode error on python 3
#46: tsa.stattools.acf confint needs checking and tests
#957: ARMA start estimate with numpy master
#62: GLSAR incorrect initial condition in whiten
#1021: from_formula() throws error - problem installing
#911: noise in stats.power tests
#472: Update roadmap for 0.5
#238: release 0.5
#1006: update nbconvert to IPython 1.0
#1038: DataFrame with integer names not handled in ARIMA
#1036: Series no longer inherits from ndarray
#1028: Test fail with windows and Anaconda - Low priority
#676: acorr_breush_godfrey undefined nlags
#922: lowess returns inconsistent with option
#425: no bse in robust with norm=TrimmedMean
#1025: add_constant incorrectly detects constant column
