Prediction (out of sample)¶
[1]:
%matplotlib inline
[2]:
import numpy as np
import matplotlib.pyplot as plt
import statsmodels.api as sm
plt.rc("figure", figsize=(16, 8))
plt.rc("font", size=14)
Artificial data¶
[3]:
nsample = 50
sig = 0.25
x1 = np.linspace(0, 20, nsample)
X = np.column_stack((x1, np.sin(x1), (x1 - 5) ** 2))
X = sm.add_constant(X)
beta = [5.0, 0.5, 0.5, -0.02]
y_true = np.dot(X, beta)
y = y_true + sig * np.random.normal(size=nsample)
Estimation¶
[4]:
olsmod = sm.OLS(y, X)
olsres = olsmod.fit()
print(olsres.summary())
OLS Regression Results
==============================================================================
Dep. Variable: y R-squared: 0.979
Model: OLS Adj. R-squared: 0.977
Method: Least Squares F-statistic: 704.2
Date: Tue, 13 Jan 2026 Prob (F-statistic): 1.96e-38
Time: 23:44:26 Log-Likelihood: -5.8313
No. Observations: 50 AIC: 19.66
Df Residuals: 46 BIC: 27.31
Df Model: 3
Covariance Type: nonrobust
==============================================================================
coef std err t P>|t| [0.025 0.975]
------------------------------------------------------------------------------
const 4.9996 0.097 51.745 0.000 4.805 5.194
x1 0.4970 0.015 33.354 0.000 0.467 0.527
x2 0.4798 0.059 8.191 0.000 0.362 0.598
x3 -0.0197 0.001 -15.029 0.000 -0.022 -0.017
==============================================================================
Omnibus: 1.412 Durbin-Watson: 2.161
Prob(Omnibus): 0.494 Jarque-Bera (JB): 1.334
Skew: -0.377 Prob(JB): 0.513
Kurtosis: 2.732 Cond. No. 221.
==============================================================================
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
In-sample prediction¶
[5]:
ypred = olsres.predict(X)
print(ypred)
[ 4.50807441 4.97837129 5.41082576 5.77928791 6.0670452 6.26956828
6.39525513 6.46405134 6.50417307 6.54747122 6.62419844 6.75803868
6.96221598 7.23732183 7.57121796 7.94103064 8.31690875 8.66692826
8.96233723 9.18227972 9.31722228 9.37052036 9.35786696 9.30471433
9.24209225 9.20150989 9.209779 9.28460875 9.43169622 9.64379035
9.90188328 10.17833329 10.44140624 10.66048944 10.81112118 10.87900976
10.86238152 10.77227234 10.63071437 10.46711628 10.3134315 10.1989083
10.14528446 10.16321576 10.25052528 10.39256107 10.56460343 10.73592626
10.87484529 10.95392372]
Create a new sample of explanatory variables Xnew, predict and plot¶
[6]:
x1n = np.linspace(20.5, 25, 10)
Xnew = np.column_stack((x1n, np.sin(x1n), (x1n - 5) ** 2))
Xnew = sm.add_constant(Xnew)
ynewpred = olsres.predict(Xnew) # predict out of sample
print(ynewpred)
[10.94282636 10.80479279 10.55863984 10.24724868 9.92706603 9.65428403
9.47108241 9.39530148 9.41607416 9.49648662]
Plot comparison¶
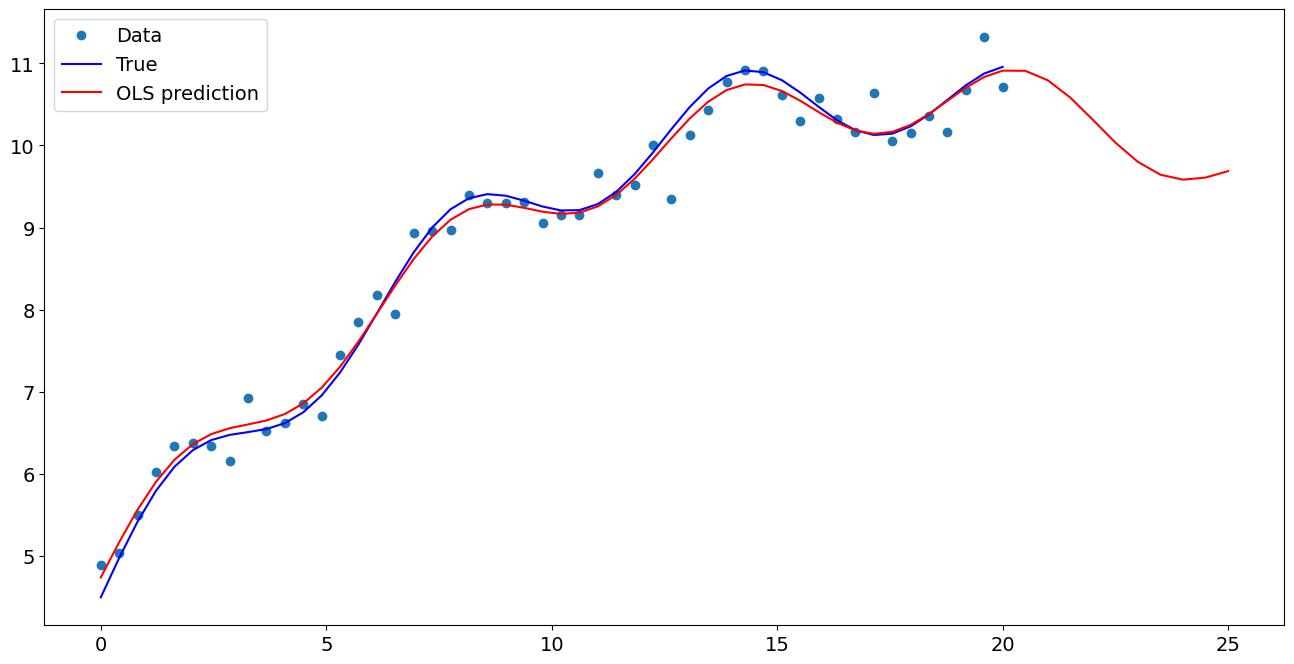
[7]:
import matplotlib.pyplot as plt
fig, ax = plt.subplots()
ax.plot(x1, y, "o", label="Data")
ax.plot(x1, y_true, "b-", label="True")
ax.plot(np.hstack((x1, x1n)), np.hstack((ypred, ynewpred)), "r", label="OLS prediction")
ax.legend(loc="best")
[7]:
<matplotlib.legend.Legend at 0x7f5724f17970>
Predicting with Formulas¶
Using formulas can make both estimation and prediction a lot easier
[8]:
from statsmodels.formula.api import ols
data = {"x1": x1, "y": y}
res = ols("y ~ x1 + np.sin(x1) + I((x1-5)**2)", data=data).fit()
We use the I to indicate use of the Identity transform. Ie., we do not want any expansion magic from using **2
[9]:
res.params
[9]:
Intercept 4.999640
x1 0.497016
np.sin(x1) 0.479823
I((x1 - 5) ** 2) -0.019663
dtype: float64
Now we only have to pass the single variable and we get the transformed right-hand side variables automatically
[10]:
res.predict(exog=dict(x1=x1n))
[10]:
0 10.942826
1 10.804793
2 10.558640
3 10.247249
4 9.927066
5 9.654284
6 9.471082
7 9.395301
8 9.416074
9 9.496487
dtype: float64